05 Reading and writing data
question Questionsobjectives Objectives
- How to import your data into RStudio?
- How to write a variable to a file?
- learn how to read text files on your computer using R
- learn how to write variables in R to a file on your computer
time Time estimation: 30 minutes
Reading and writing files
Reading files
Entering data in R can be done by typing the values when you create a variable. In most cases, however, you will have a file with data that was created by an instrument in your lab. How to import such a file into R?
There is a manual available in the R documentation called R Data Import/Export. It’s accessible using help.start() and covers in detail the functionality R has to import and export data. Reading this is highly recommended. This manual covers importing data from spreadsheets, text files, and networks.
Reading text files
Most instruments put out data in text format: tab-delimited text (.txt) or comma-separated value files (.csv). Both can be easily opened in R.
The most convenient method to import data into R is to use the read functions, like read.table(). These functions can read data in a text file. In Notepad you can save such a file as a regular text file (extension .txt). Many spreadsheet programs can save data in this format. Reading means opening the file and storing its content into a data frame.
read.table(file,header=FALSE,sep="",dec=?.?,skip=0,comment.char="#")
This function has a long list of arguments, the most important ones are:
- file: path on your computer to the file e.g. D:/trainingen/Hormone.csv If it is stored in the working directory, you can simply use its name. You can also use file=file.choose() to browse to the file and select it. File can be replaced by a url to load a file with data from the internet.
- header: does the first line of the file contain column names?
- dec: symbol used as decimal separator
- sep symbol used as column separator, default is a whitespace or tab
- skip: number of lines to skip in the file before starting to read data
- comment.char: symbol to define lines that must be ignored during reading
See the documentation for an overview of all the arguments. The output of every read function is a data frame.
There are functions to read specific file formats like .csv or tab-delimited .txt files. In the documentation of read.table() you see that these functions are called read.csv() and read.delim(). Both functions call read.table(), but with a bunch of arguments already set. Specifically they set up sep to be a tab or a comma, and they set header=TRUE.
read.delim(file,header=TRUE,sep="\t")
On the documentation page, you see that these functions each have two variants that have different default settings for the arguments they take:
read.csv( file,header=TRUE,sep= ",",dec=".", ...)
read.csv2( file,header=TRUE,sep= ";",dec=",", ...)
read.delim( file,header=TRUE,sep="\t",dec=".", ...)
read.delim2(file,header=TRUE,sep="\t",dec=",", ...)
Originally the CSV format was designed to hold data values separated by commas. In .csv files that are made on American computers this is the case. However, in Europe the comma was already used as a decimal separator. This is why .csv files that are made on a European computer use the semicolon as a separator.
For instance, the file below contains a header row and three columns, separated by semicolons. It uses the comma as decimal separator.
Patient;Drug;Hormone
1;A;58,6
2;A;57,1
3;B;40,6
Obviously, the file is a European CSV file, to open it use read.csv2()
Reading Excel files
To import Excel files via a command the easiest way is to let Excel save the file in .csv or tab delimited text format and use the read functions.
An easy way to import Excel files is to use the RStudio interface although I prefer to use commands. To use the interface go to the Environment tab and click the Import Dataset button.
RStudio can import 3 categories of files: text files, Excel files and files generated by other statistical software. To read .xls or .xlsx files select From Excel.
A dialog opens with options on the import. You can import data from your computer (Browse) or from the internet (provide a url and click Update). Click Browse, locate the Excel file and click Open.
The Data Preview section shows what the data will look like in R.
The Import Options section allows you to specify the import parameters.
- Name: name of the data frame that will hold the imported data. The default is the name of the file that you are opening.
- Skip: number of rows at the top of the file to skip during import. Some data formats contain a number of header rows with general info like parameter settings, sample names etc. These rows are followed by the actual data. Skip allows you to skip over the header rows and import the actual data.
- If the first row of the file contains column names, select First Row as Names
- Open data viewer shows the data in the script editor upon import
Click Import.
Behind the scenes RStudio uses the readxl package that comes with the tidyverse package. You can also use the functions of this package directly in commands.
Compared to other packages for reading Excel files (gdata, xlsx, xlsReadWrite) readxl has no external dependencies, so it?s easy to install and use on all operating systems. It supports the .xls format and the .xlsx format. The easiest way to install it from CRAN is to install the whole tidyverse package but you have to load readxl explicitly, since it is not a core tidyverse package.
Once imported into RStudio the data is stored in a data frame and you can use it as input of commands. The data frame appears in the list of Data in the Environment tab.
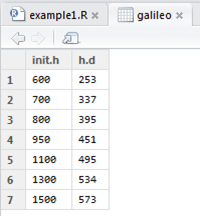
If you want to view the data frame you can click its name in the Environment tab and it will appear in a separate tab in the script editor.
hands_on Hands-on: Demo
From the demo script run the Reading files section
hands_on Hands-on: Exercise 17a
- Import the file GeneEx.csv into a data frame called GeneEx
- Rename the two last columns Ct1 and Ct2
- Create a new column containing the average Ct: (Ct1+Ct2)/2
solution Solution
GeneEx <- read.csv2("Rdata/GeneEx.csv") colnames(GeneEx)[c(3,4)] <- c("Ct1","Ct2") GeneEx$Average_Ct <- (GeneEx$Ct1 + GeneEx$Ct2)/2question Question
Which of these 2 commands will work ?
GeneEx <- read.csv2("Rdata/GeneEx") GeneEx <- read.csv2("http://data.bits.vib.be/pub/trainingen/RIntro/GeneEx.csv")question Question
Which of these 2 commands will work ?
names(GeneEx[c(3,4)]) <- c("Ct11","Ct21") names(GeneEx)[3:4] <- c("Ct11","Ct21")question Question
What’s the difference in result between these 2 commands ?
GeneEx$Average_Ct2 <- (GeneEx$Ct1+GeneEx[4])/2 GeneEx[5] <- (GeneEx[3]+GeneEx[4])/2question Question
Can you use sum() instead of + ?
sum(GeneEx$Ct1,GeneEx$Ct2) (GeneEx$Ct1+GeneEx$Ct2)question Question
Can you use mean() instead of +/2 ?
mean(GeneEx$Ct1,GeneEx$Ct2) mean(GeneEx$Ct1)
Reading other files
Also of note is an R package called foreign. This package contains functionality for importing data into R that is formatted by most other statistical software packages, including SAS, SPSS, STRATA and others.
Writing files
Reversely, to write a data frame to a file you can use the generic function:
write.table(x,file=?name.txt?,quote=TRUE,row.names=TRUE,col.names=TRUE)
This function has a long list of arguments, the most important ones are:
- x: data frame to be written to a file
- file: name or full path of the file e.g. D:/trainingen/Hormone.csv
- quote: if TRUE, strings, row and column names will be surrounded by double quotes. If FALSE, nothing is quoted.
- sep: column separator
- row.names: boolean indicating whether the row names of x are to be written or a character vector of row names to be written
- col.names: boolean indicating whether the column names of x are to be written or a character vector of column names to be written
- append=FALSE: if TRUE x is added to the file defined by file
- eol = ?\n?: end-of-line character, default ?\n? represents an enter
- na=?NA?: string to use for missing values in the data
- dec=?.?: decimal separator
See the help file for a full overview of all arguments.
To specifically write .csv files use write.csv() or write.csv2(). See the help file for a description of the difference between them.
Excel can read .csv files but if you really want to write .xls or .xlsx files use the openxlsx package.
hands_on Hands-on: Exercise 17b
- Read the file RNASeqDE.txt into a data frame called DE. It contains the differentially expressed genes from an RNA-Seq experiment.
- Split the table into a table of upregulated genes (log2foldchange > 0) and a table of downregulated genes and store them in data frames called up and down.
- How many up- and downregulated genes are there?
- What is the gene with the highest log2 fold change?
- What is the data of the gene with the lowest adjusted p-value (= padj)?
- Write the Ensembl IDs (= row names) of the upregulated genes to a file called up.txt. You will use this file for functional enrichment analysis using online tools like ToppGene,EnrichR? These tools want a file with only Ensembl IDs as input (one per line, no double quotes, no column headers, no row names).
solution Solution
DE <- read.table("Rdata/RNASeqDE.txt",header=TRUE) up <- DE[DE$log2FoldChange > 0,] down <- DE[DE$log2FoldChange < 0,] nrow(up) nrow(down) rownames(up[which.max(up$log2FoldChange),]) DE[which.min(DE$padj),] write.table(rownames(up),file="up.txt",quote=FALSE,col.names=FALSE,row.names=FALSE)question Question
Which of the following 2 commands will not work properly ?
DE <- read.table("Rdata/RNASeqDE.txt") file <- file.choose() DE <- read.table(file,header=TRUE)question Question
Will the following command work ?
up <- subset(DE,log2FoldChange > 0)question Question
What’s the difference between these 2 commands ?
which.max(up$log2FoldChange) max(up$log2FoldChange)question Question
Will this command write Ensembl IDs and log fold changes ?
toprint <- as.data.frame(up$log2FoldChange) write.table(toprint,file="up.txt",quote=FALSE,col.names=FALSE)
hands_on Hands-on: Extra exercise 17c
Which type of files are imported by read.delim ?
solution Solution
Check the documentation and look at the default for sep
hands_on Hands-on: Extra exercise 17d
- Read the file ALLphenoData.tsv into a variable called pdata using one of the read functions
- What type of data structure is pdata ?
- What are the names of the columns of pdata ?
- How many rows and columns are in pdata ?
solution Solution
pdata <- read.delim("Rdata/ALLphenoData.tsv") class(pdata) colnames(pdata) dim(pdata)
keypoints Key points
- We showed the different functions in R to text read files
- We showed how to write the values of a variable to a text file
